Submitted in partial fulfilment of the requirements for the degree of BSc. in Information Technology (Software Development) at the University of Malta.
Introduction
Diabetes is a global health problem that impacts around 463 million people [1]. Diabetic patients are at higher risk of critical glycaemic events. These events can be mitigated through timely intervention; however, these preventive actions take time to act. Hence the prediction of blood
glucose levels using machine learning (ML) is of interest, to provide the patient with predictions on future blood glucose levels.
The performance of ML algorithms is dependent on their hyperparameter configuration. The manual selection of hyperparameters can often be challenging due to the large search spaces. Hyperparameter optimisation may be used to algorithmically select and tune the hyperparameters of a ML model. Metaheuristic optimisers use intelligent ways of exploring the search space to find near optimal solutions using exploration and exploitation mechanisms among other factors
Aims
The aim of this study was to improve the performance of ML models on the ohioT1DM dataset. By applying more than one metaheuristic optimiser for the task of finding candidate combinations of
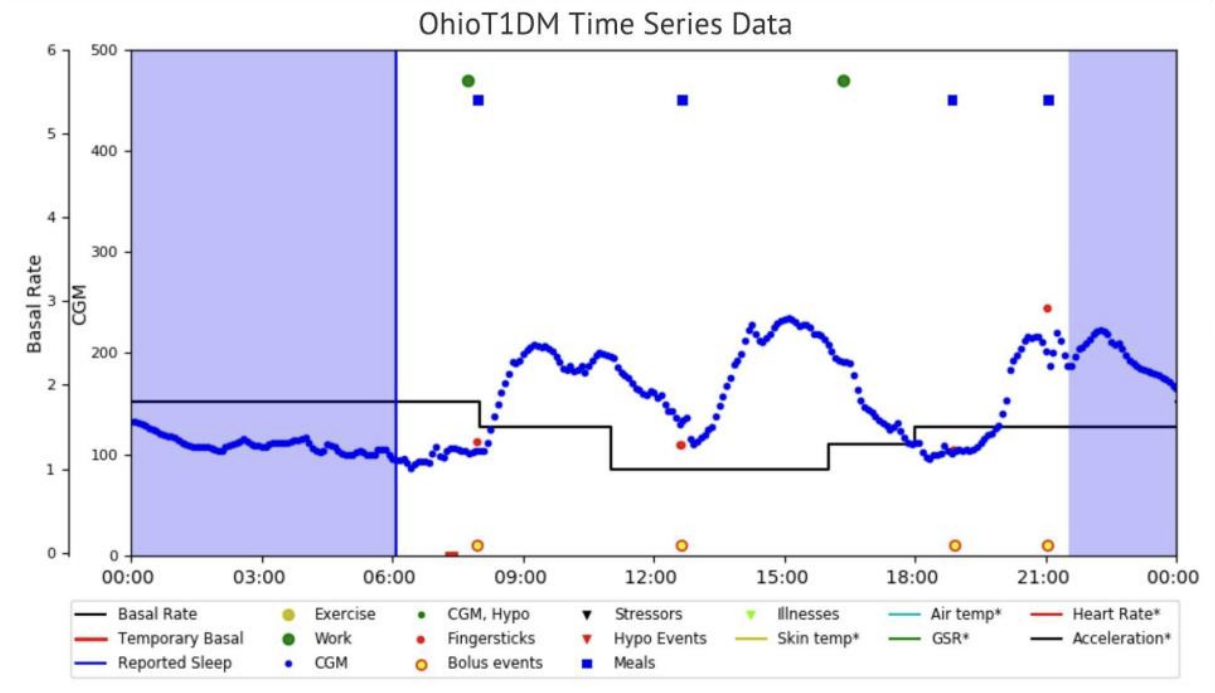
hyperparameters. The ohioT1DM dataset is a publicly available dataset, consisting of physiological time series data of several Type-1 diabetes patients collected over eight weeks [2]. The dataset contains thousands of rows and multiple columns, obtained from wearable sensors and a continuous glucose monitor (CGM). The study was also looking at physiological measures other than continuous blood glucose readings.
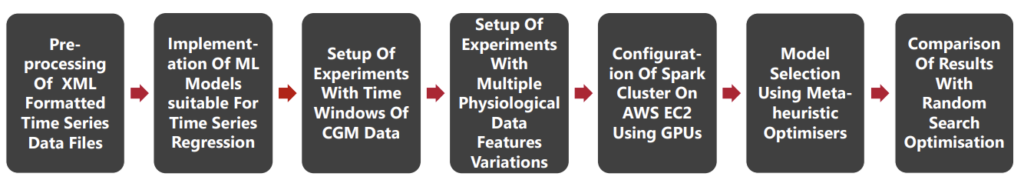
Methadology
The following are the main steps of the methadology in order to preprocess the data, fit the models, optimise the models and evaluation of results.
Genetic algorithm
The implementation of a genetic algorithm in Python involves several key steps. First, a population of potential solutions, known as individuals is initialized, typically with random values. Each individual represents a possible solution to the problem. The fitness of each individual is then evaluated using a predefined fitness function that measures how well the solution meets the desired objectives. Next, individuals are selected for reproduction based on their fitness, with better-performing individuals having a higher chance of being chosen.
These selected individuals undergo crossover to produce offspring, which involves exchanging segments of their genetic information to create new individuals. Mutation is then applied to introduce small random changes to some of the offspring, ensuring genetic diversity within the population. The new generation of individuals replaces the old population, and the process repeats for a specified number of generations or until a convergence criterion is met. Throughout these steps, the algorithm evolves the population towards increasingly optimal solutions.
The following is my implementation of the genetic algorithm in python.
Experimentation and Results
The experimentation included training an MLP, RNN and XGBoost using metaheuristic optimisers, the genetic algorithm and particle swarm optimisation. Such experiments were varied by the inclusion of different subsets of the physiological features as the input data to the models. Due to the random element in the optimisation processes, the optimisation runs were performed five times each in order to obtain the average improvement and thus a better estimate of the true performance. Comparisons were performed between different optimisation approaches. A closer look was taken at the rate of improvement by looking at the average improvement and standard deviation from one generation to the next. A consequence of running the experimentation multiple runs is a higher computational load, hence the workload was distributed using Spark among a number of cloud instances with GPU resources for faster training.
Conclusion
In this study the use of metaheuristic approaches to optimise machine learner performance on blood glucose levels prediction together with the use of other physiological data and alternative means to increase computational power were explored. Initial experiments indicate that a metaheuristic approach to hyperparameter optimisation in this context may produce better
results than random search given sufficient generations. It is noted that using such techniques significantly increases the computational cost, as the models must be retrained multiple times with different configurations.
Possible future work is exploring the use of adaptive metaheuristic optimisers and the tuning of parameters that impact the exploration and exploitation aspect of the metaheuristic algorithms. Due to the prevalence of Type-2 diabetes in Malta [2], techniques implemented in this paper may be adapted for blood glucose levels prediction using physiological time series data obtained from Maltese Type-2 diabetes patients.